Abstract
INTRODUCTION
Evolving techniques have made possible the direct detection, physical isolation, and study of AML minimal residual disease (MRD) after treatment. This could allow for better identification of therapeutic vulnerabilities in AML. Prior studies have focused on cells that initiate leukemia in mouse models, known as leukemia-initiating cells (LIC), generally with a foundational CD34+CD38- immunophenotype. LIC are typically derived from diagnostic samples of untreated patients. Such stem-like cells do not necessarily represent the residual fraction of AML after treatment. Relapse may originate from non-LIC, and the presence of phenotypically and molecularly defined MRD is now firmly established as a critical prognostic factor for patients. High-risk AML is characterized by relapse, despite morphologic complete remission with initial therapy in most cases. RNA-sequencing was performed on pre- and post-treatment AML subpopulations, including MRD, from high-risk patients, to determine differences in gene expression.
METHODS
Matched primary AML samples were collected from marrow and peripheral blood of patients with high-risk AML (including patients with unfavorable karyotype and/or TP53 mutation) at diagnosis and after treatment. Mononuclear cells were flow-sorted for bulk (CD45dim) and LIC (Lin-CD34+CD38-CD123+) from diagnostic samples. Post-treatment samples were sorted for bulk mononuclear cells (MNC) and MRD, based on difference-from-normal/MRD immunophenotype specific for each patient as determined from established 20-marker clinical flow cytometry analysis. RNA was isolated using low-input methodology, and RNA-sequencing was performed using Illumina HiSeq 2000. Gene expression was assessed using GO-Elite, and differences between patients and subpopulations were assessed using rank product method.
RESULTS
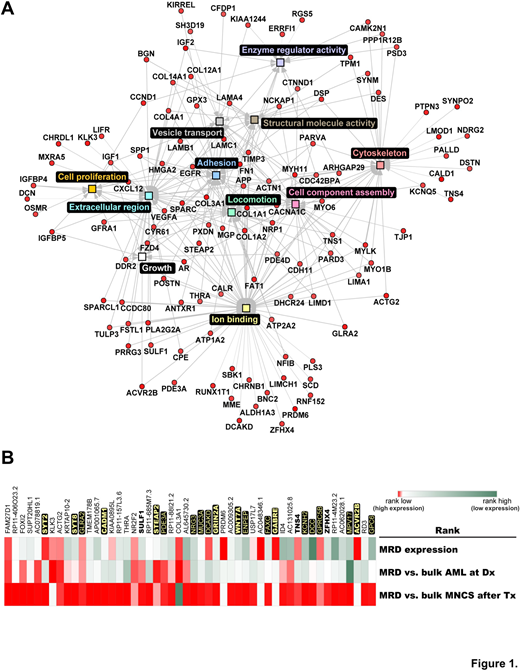
Gene expression in MRD was analyzed by RNA-sequencing in comparison to diagnostic samples in eight patients with high-risk AML. Four patients had unfavorable karyotype, including two with TP53 mutations. Patients had additional high-risk features, such as FLT3-ITD or RUNX1 mutations, or secondary/therapy-related AML. Treatment consisted of chemotherapy (6/8) or hypomethylating agents (2/8), with or without other targeted drugs. Residual leukemia was detected in post-treatment samples in all study patients. Significant differences in gene expression were detected between MRD and other sorted populations, including diagnostic bulk AML and LIC. Relevant MRD pathways included those with strong interactions with the microenvironment. Anti-apoptotic mechanisms, cytoskeletal, and cell adhesion related genes, WNT/beta-catenin signaling, and TGFbeta signaling ranked among the most relevant processes in AML MRD subpopulations (Figure 1A, GO-Elite interactome of highly expressed genes in AML MRD). To identify potentially critical and unique MRD-specific genes, rank product method was applied using 1) the most highly expressed genes in AML MRD, 2) the most differential expressed genes between MRD and bulk AML at diagnosis, and 3) the most differentially expressed genes between MRD and bulk MNC after treatment. Among the top 50 scoring genes using this approach (Figure 1B), 16 genes were among the top 5% of genes expressed in MRD among all patients and 20 genes have cell surface gene-products (shown in yellow). Several potential leukemia- and cancer-related genes of interest were identified (shown in bold).
CONCLUSIONS
Key differences exist between the gene expression profiles of post-treatment MRD from high-risk AML patients, in comparison to other populations and subpopulations of sorted cells before and after treatment. The highlighted differences suggest that MRD relies on specific intrinsic gene expression changes and microenvironmental interactions, and therefore may be targetable after elimination of bulk AML with initial therapy. Accessible surfacesome targets are among top hits.
Konopleva:cellectis: Research Funding; Immunogen: Research Funding; abbvie: Research Funding; Stemline Therapeutics: Research Funding. Andreeff:Astra Zeneca: Research Funding; Amgen: Consultancy, Research Funding; Jazz Pharma: Consultancy; Celgene: Consultancy; Reata: Equity Ownership; SentiBio: Equity Ownership; Aptose: Equity Ownership, Membership on an entity's Board of Directors or advisory committees; United Therapeutics: Patents & Royalties: GD2 inhibition in breast cancer ; Eutropics: Equity Ownership, Membership on an entity's Board of Directors or advisory committees; Oncolyze: Equity Ownership; Daiichi-Sankyo: Consultancy, Patents & Royalties: MDM2 inhibitor activity patent, Research Funding; Oncoceutics: Equity Ownership, Membership on an entity's Board of Directors or advisory committees.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal